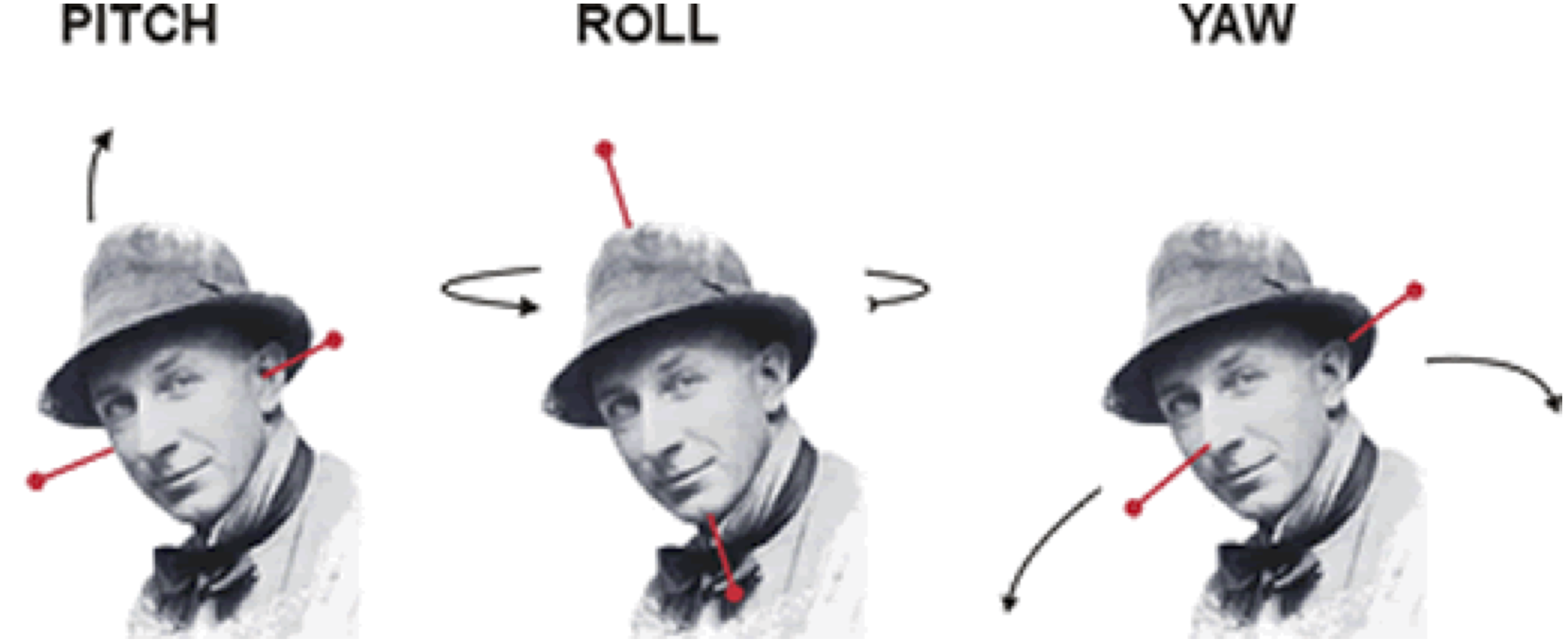
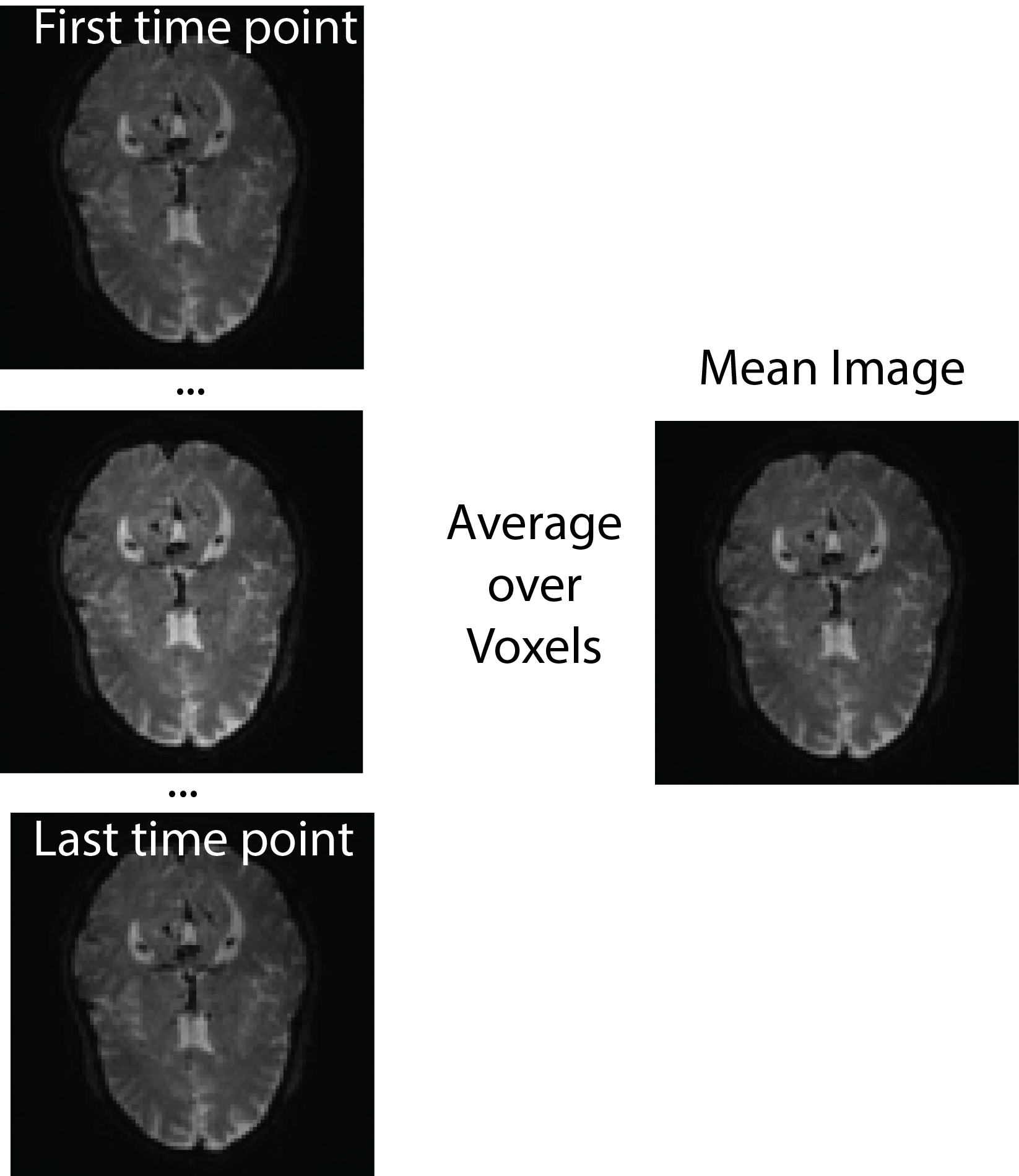
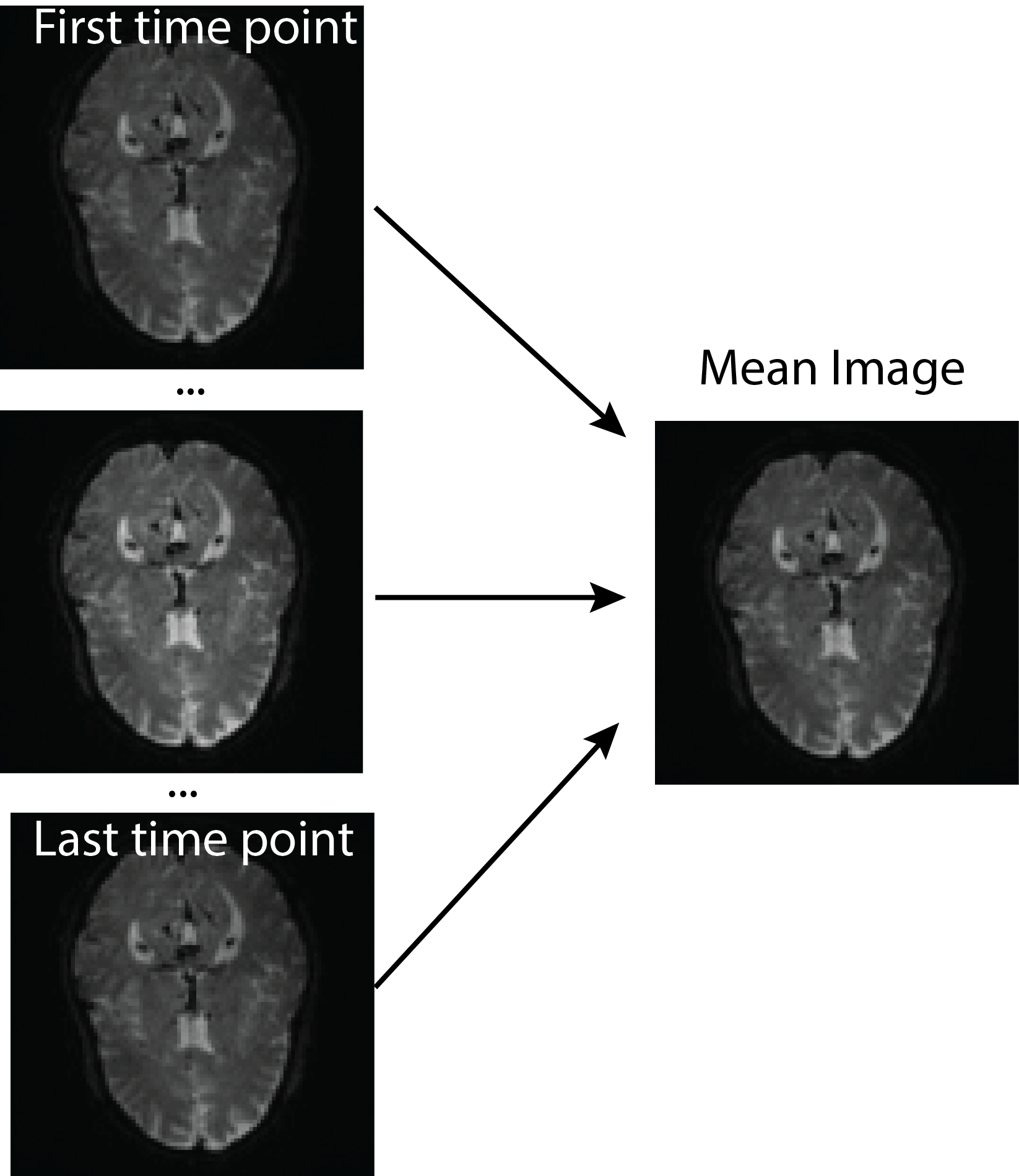
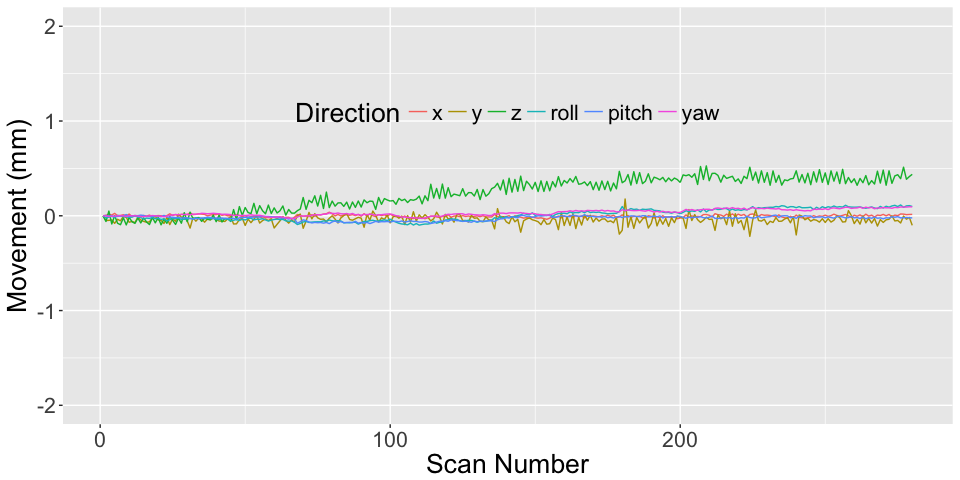
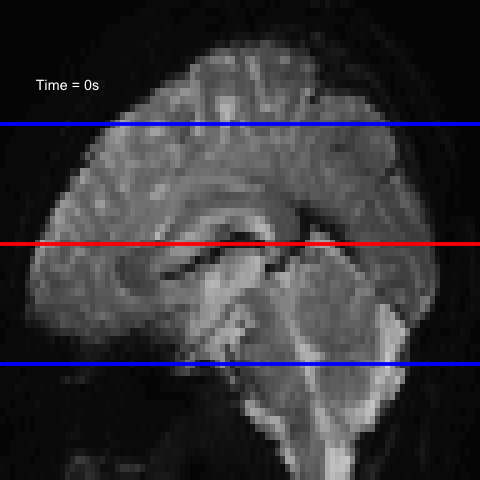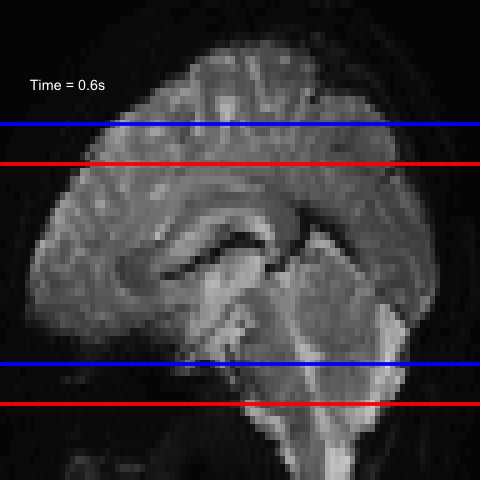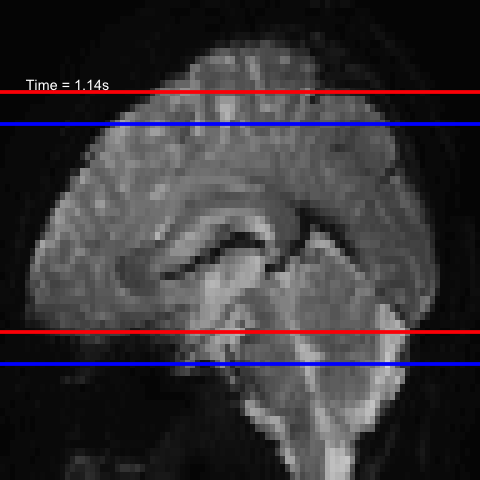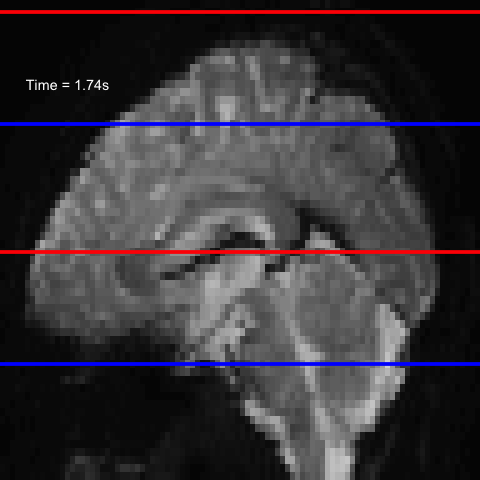
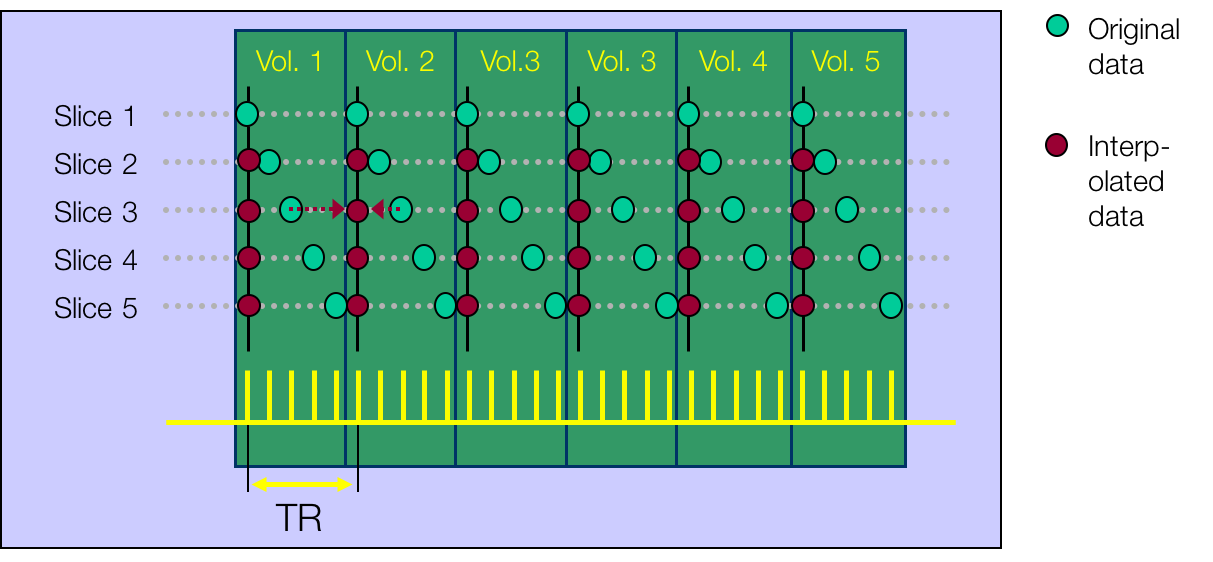
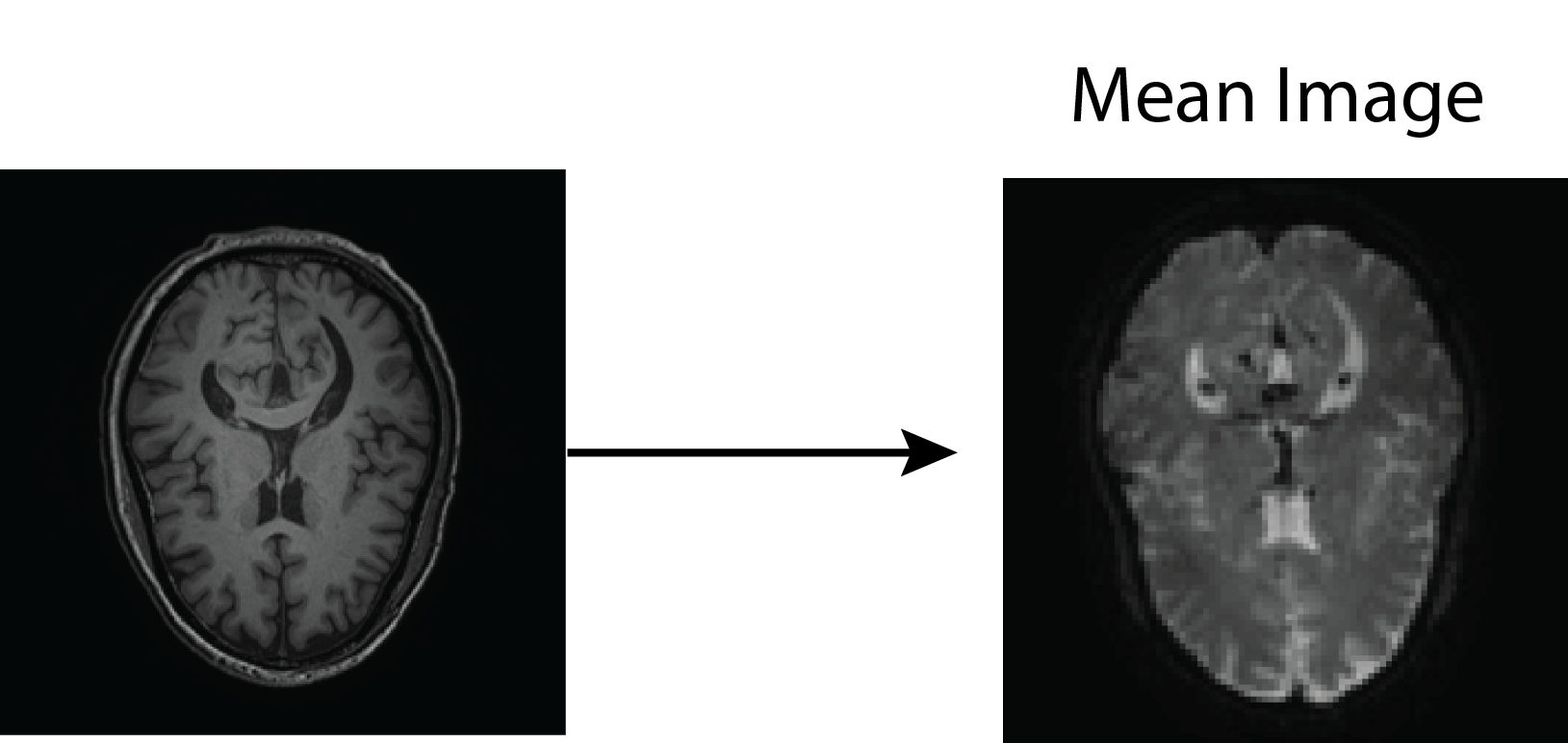
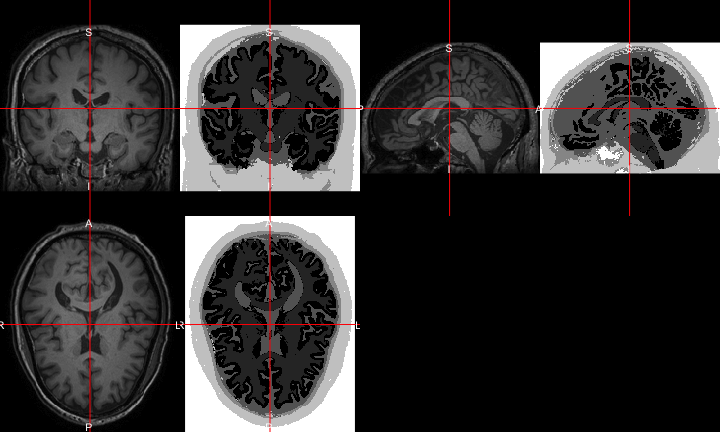
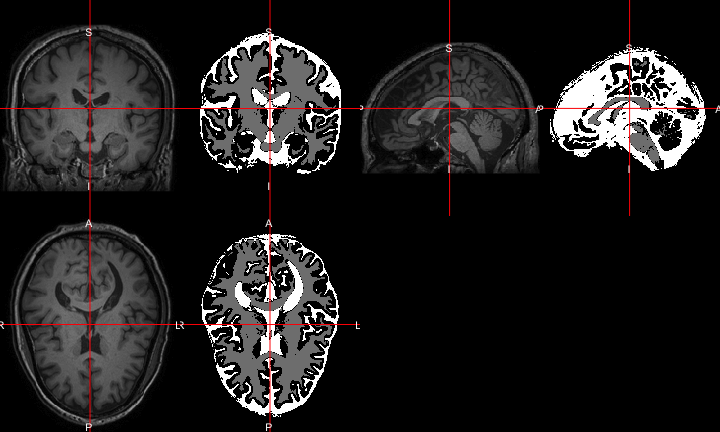
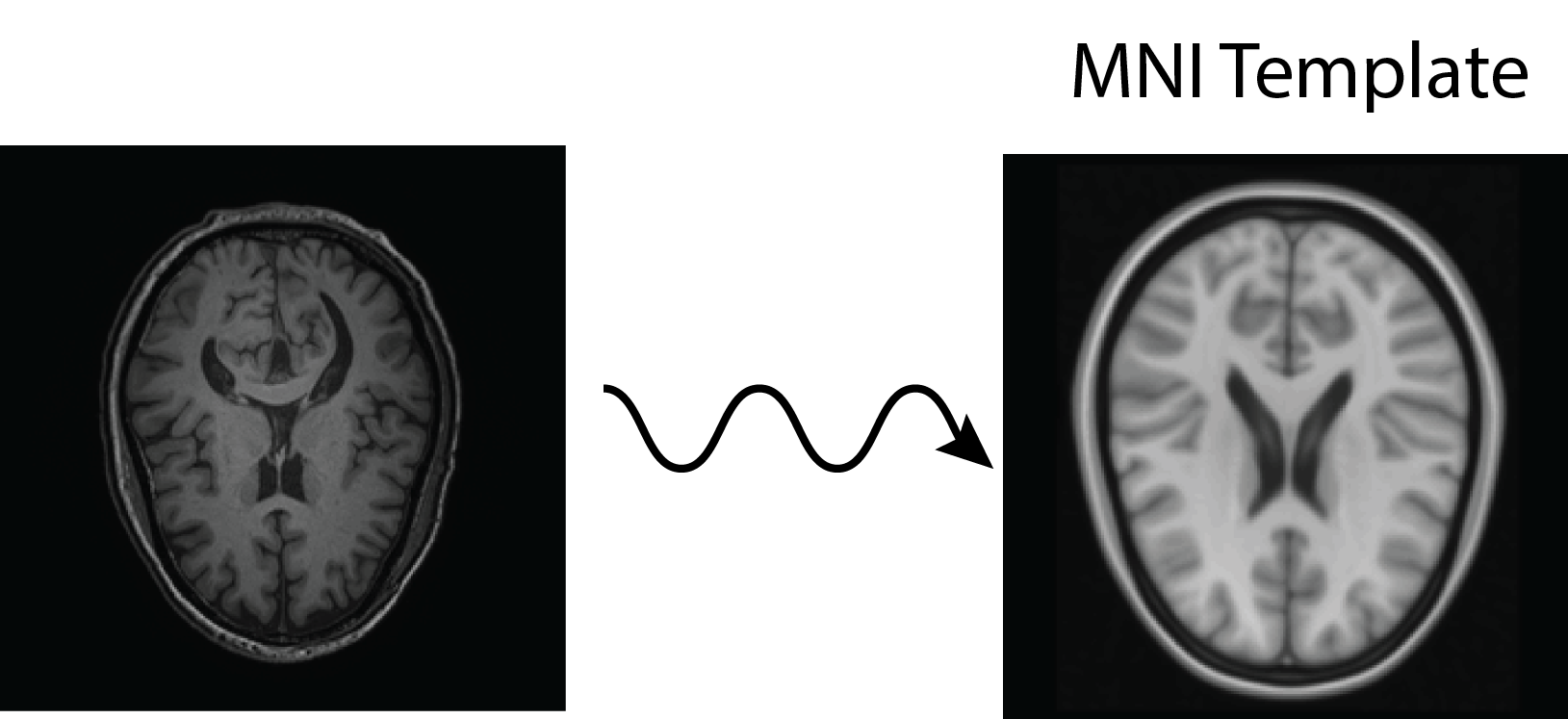
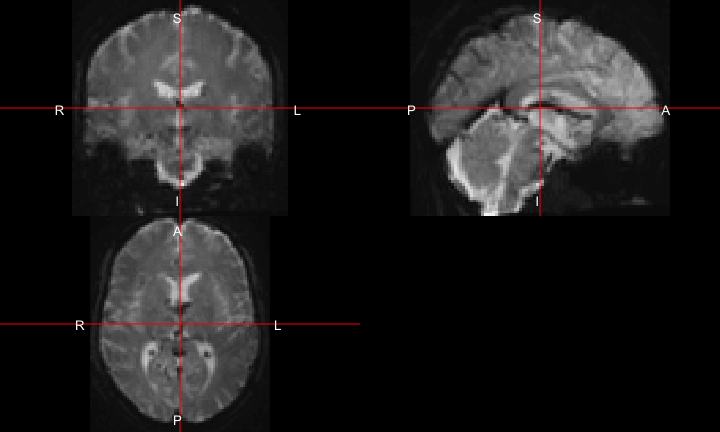
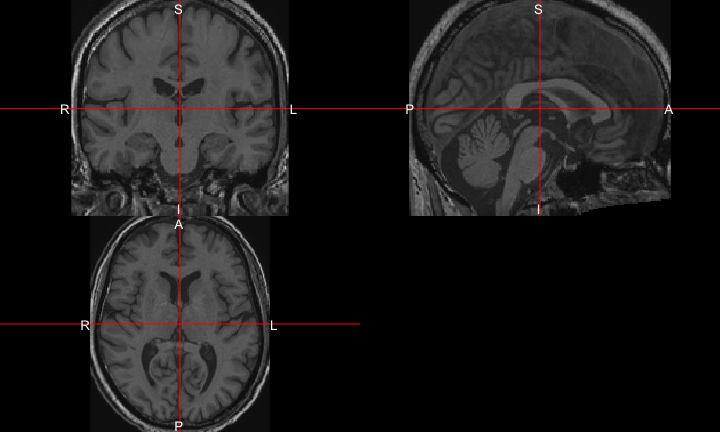
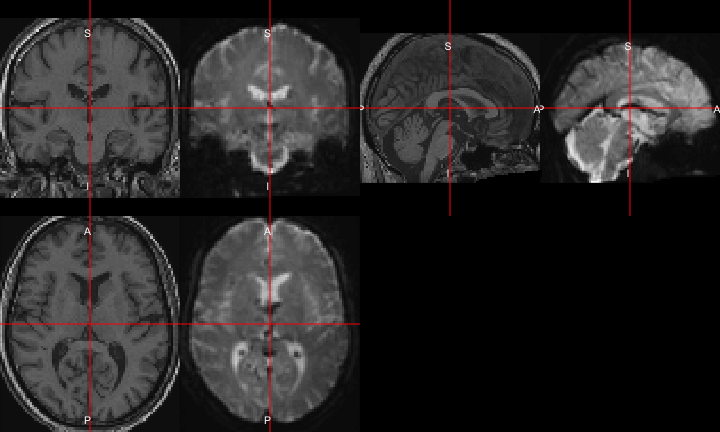
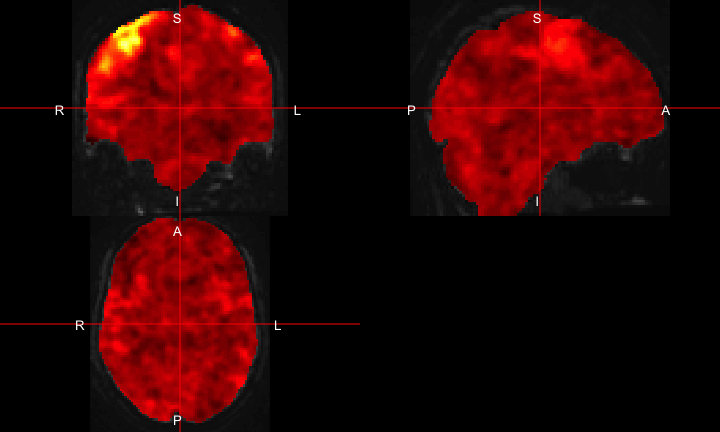
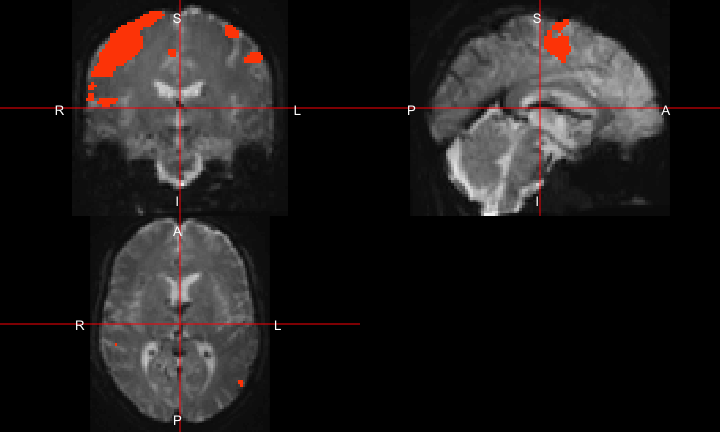
All of this is using the Statistical Parametric Mapping (SPM) (Penny et al. 2011) software version 12:
- requies MATLAB (for this tutorial)
- All called through
spm12rpackage - All code is found at https://github.com/muschellij2/talks/tree/master/fmri_task_processing